A new approach integrating skeleton graph optimization and fractal self-similarity has significantly improved the accuracy of three-dimensional (3D) tree modeling. This innovative method, developed by researchers led by Zhenyang Hui at the East China University of Technology, addresses common errors found in existing models, such as misconnected branches and gaps due to incomplete data scans. The findings were published on June 1, 2025, in the journal Plant Phenomics.
The method was rigorously tested on 29 trees from diverse tropical forest sites in Peru, Indonesia, and Guyana. Results indicated a remarkable concordance correlation coefficient of 0.994, surpassing widely used models like TreeQSM and AdQSM. Accurate 3D models are crucial for calculating parameters such as diameter at breast height (DBH), above-ground biomass, and wood volume, which are essential for assessing carbon stocks and understanding forest structure.
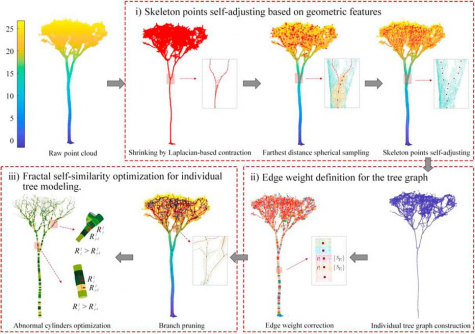
LiDAR (Light Detection and Ranging) technology is increasingly utilized for capturing point cloud data necessary for these models. Traditional methods, however, often produce fragmented representations of trees and fail to effectively handle incomplete canopy data. Such limitations hinder their reliability in large-scale forest monitoring and biodiversity assessments.
To tackle these challenges, Hui’s team developed a new 3D modeling method, termed SfQSM. Utilizing LiDAR datasets collected from three tropical forest locations, the researchers employed a Riegl VZ-400 terrestrial laser scanner that operates at 1550 nm, capturing high-resolution point clouds with 1 cm precision. To validate the model’s accuracy, all trees were destructively harvested, allowing for the calculation of stem, buttress, and large branch volumes using standard forestry formulas.
The study revealed that SfQSM delivered highly accurate volume estimates, with deviations from the harvested volumes typically falling between −1 m3 and 1 m3. Smaller-diameter trees from Indonesia showed lower deviations, while larger trees from Peru exhibited greater discrepancies, indicating that tree size plays a significant role in modeling precision. Evaluation metrics indicated a mean deviation of 0.162 m3, a root mean square error of 1.023 m3, and relative errors as low as 0.01% and 0.09%. In contrast, TreeQSM and AdQSM demonstrated significantly higher error rates, with TreeQSM’s errors being more than double those of SfQSM and AdQSM’s deviations exceeding SfQSM’s by over thirtyfold.
Visual analyses further underscored the robustness of the new method. While TreeQSM often resulted in fragmented trunks and AdQSM generated overfitted or non-existent branches, SfQSM consistently produced continuous and realistic models aligned with the point clouds. These results not only confirm SfQSM’s superiority in statistical accuracy but also illustrate its capacity for high-fidelity structural reconstructions.
The implications of this research are substantial. By generating precise individual tree models, the method provides a reliable foundation for biodiversity assessments, species classification, and habitat analysis. It also enhances the estimation of tree volume and biomass, which is critical for determining carbon stocks and tracking forests’ contributions to the global carbon cycle.
In addition, accurate 3D reconstructions can improve virtual ecological landscapes, aiding in the development of sustainable forestry practices, reforestation initiatives, and climate change mitigation strategies. The funding for this research was provided by various sources, including the National Key Laboratory of Uranium Resources Exploration-Mining and Nuclear Remote Sensing and the National Natural Science Foundation of China.
This study represents a significant advancement in the field of ecological modeling, offering vital tools for forestry, carbon monitoring, and ecological conservation.