A research team led by doctoral candidates at the University of Maine has developed a groundbreaking artificial intelligence (AI) tool designed to improve the accuracy and speed of breast cancer diagnosis. The system, named the Context-Guided Segmentation Network (CGS-Net), mimics the methods that human pathologists use to analyze tissue samples, potentially preventing delays in diagnosis that can be life-threatening.
Spearheaded by Jeremy Juybari, pursuing a Ph.D. in electrical and computer engineering, and Josh Hamilton, a Ph.D. candidate in biomedical engineering, the tool employs a sophisticated deep learning architecture. This innovation is aimed at interpreting microscopic images of tissue with greater precision than traditional AI models, which often fall short in accuracy.
Breast cancer remains a critical health issue, being the second leading cause of cancer-related deaths among women worldwide. Statistics indicate that one in eight women will be diagnosed with breast cancer in their lifetime. Diagnosis typically relies on the meticulous inspection of chemically stained tissue samples, a process that demands considerable expertise and time. Compounding the challenge, two-thirds of the world’s pathologists are concentrated in just ten countries, resulting in significant diagnostic delays in many regions. For instance, in India, approximately 70% of cancer fatalities are attributed to treatable risk factors exacerbated by limited access to timely diagnostics.
Juybari explained the innovation behind CGS-Net, stating, “This model integrates both detailed local tissue regions and broader contextual regions to improve the accuracy of cancer predictions in histological slides.” He noted that the research illustrates how incorporating the context surrounding tissue can significantly enhance predictive performance. This study is documented in a paper published in Scientific Reports, co-authored with faculty researchers Andre Khalil, Yifeng Zhu, and Chaofan Chen. Both the dataset and source code are publicly available, promoting transparency and collaboration within the scientific community.
How CGS-Net Functions
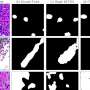
At the heart of CGS-Net is a dual-encoder model that replicates the workflow of pathologists. Traditionally, pathologists gather data by zooming in and out of images to examine tissue. In contrast, CGS-Net processes these images simultaneously. One branch of the network analyzes high-resolution patches to capture cell-level details, while the other inspects lower-resolution patches that provide a broader architectural context. This dual approach allows specialists to differentiate between normal and malignant structures effectively.
Each patch is centered on the same pixel, ensuring alignment between the two views, which are then analyzed through interconnected encoders and decoders. To validate the system’s efficacy, the research team trained CGS-Net on 383 digitized whole-slide images of lymph node tissue, determining which samples indicated breast cancer and differentiating between healthy and cancerous tissue. The results showed that the tool consistently outperformed conventional single-input models.
“The CGS-Net successfully mimicked how a pathologist examines histological samples, employing two encoders that simultaneously evaluate different levels of magnification,” the researchers highlighted in their paper.
Future Prospects and Broader Implications
While the current research focuses on binary cancer segmentation, the team anticipates broader applications. Future work will aim to incorporate multiple resolutions and expand the system’s capabilities to include multiclass tissue segmentation. There is also potential for the architecture to integrate multimodal data, such as radiology scans or molecular profiles, which could further enhance diagnostic accuracy.
Beyond its technical advancements, this project exemplifies the interdisciplinary strength of the University of Maine’s research ecosystem. It merges engineering, computing, and biomedical sciences to address global health disparities. As cancer diagnosis increasingly shifts toward digital methodologies, tools like CGS-Net are poised to augment, rather than replace, human expertise. By enabling machines to interpret images as doctors do, researchers at UMaine are paving the way for a future where early and accurate cancer detection is more accessible to all.
For more information, refer to the research paper: Jeremy Juybari et al, Context-guided segmentation for histopathologic cancer segmentation, Scientific Reports (2025). DOI: 10.1038/s41598-025-86428-7.